
Single-cell nuclear transcriptome sequencing resolves cellular heterogeneity in a single segmental compression model of chronic spinal cord injury in rabbits
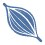
Cell number: 36,856
Timepoint: Control group, 5 days post-injury (5 DPI) group and 10 days post-injury (10 DPI) group
View cell atlas in ShinyCell: https://lifeomics.shinyapps.io/mskca_spinal_cord_001/
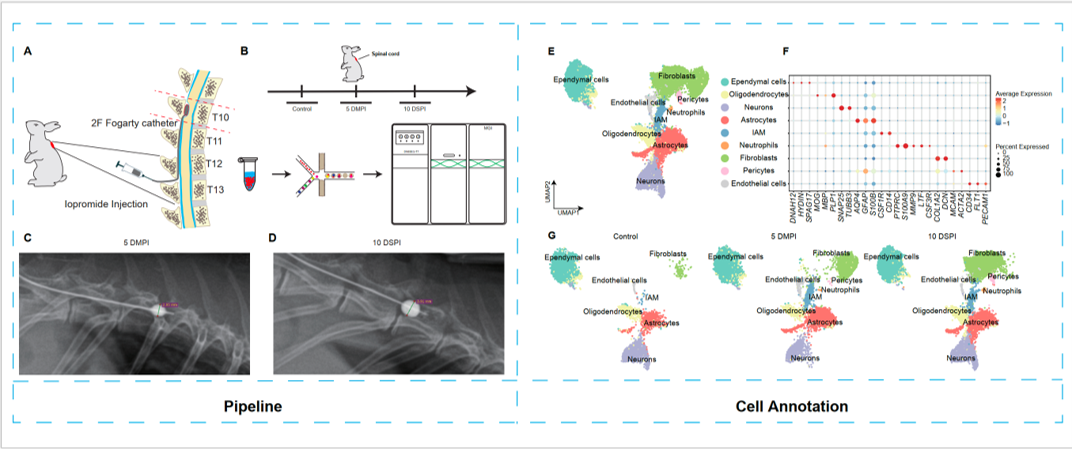
The incidence and prevalence of non-traumatic spinal cord injuries are gradually increasing with aging. Among them, spinal cord compression caused by degenerative disc diseases (DDD) and spinal tumors has gradually become the most common cause of non-traumatic spinal cord injuries. Long-term compression of the spinal cord can lead to irreversible damage to the spinal cord, with a high disability rate. Previous studies on spinal cord injury models have focused on the spinal cord injury caused by cutting/contusion. However, the nature of these injuries is transient and non-cumulative. Spinal cord injury from spinal cord compression can be either short-term or long-term and has a cumulative nature. However, the pathophysiological changes in the spinal cord after non-traumatic spinal cord injury are not fully understood. In this study, we constructed a single segmental compression model of chronic spinal cord injury in rabbits. We used snRNA-Seq to resolve the heterogeneity of different cell subsets in different stages of spinal cord compression for the first time, focusing on inflammation-related Altered biological processes in the development of spinal cord compression injury. Second, we explored the cellular heterogeneity of injury-activated microglia and macrophages (IAMs). In addition, we investigated possible interactions between neurovascular units and immune cells during different stages of spinal cord compression. Finally, we stored the spinal cord compression model's single-cell nuclear transcriptome sequencing data in the MSKCA database (www.mskca.tech/spinalcord) and performed an interactive visualization. This research will help researchers understand the changes in gene expression during the development of spinal cord compression injury and provide new insights for exploring therapeutic targets for spinal cord compression injury.